DISCUSSION
Biosynthetic natural rubber and chemosynthetic rubber have
been used by humans at a level of about 106 tons/year for
about a century. The intrinsic biodegradability of polyisoprene by
microorganisms in the environment has been known since the first
publication of Söhngen and Fol in 1914 . Many
other reports on biodegradation of polyisoprene have appeared during
the last few decades. In particular, the number of isolated
rubber-degrading microorganisms has increased during the last 10
years . The
biochemical mechanism by which the rubber backbone is cleaved is only
poorly understood, however, and until now not one enzyme involved in
rubber degradation has been isolated.
In this study we purified an extracellular protein from
polyisoprene-grown Xanthomonas sp. cultures (RoxA) that has
the ability to cleave the carbon backbone of polyisoprene in vitro,
and we characterized its biochemical properties. A low-molecular-mass
compound derived from a three-isoprene-unit backbone
(12-oxo-4,8-dimethyltrideca-4,8-diene-1-al; m/z 236) was
identified as the major degradation end product of in vitro rubber
degradation by purified RoxA. Four additional minor products that
differed by a mass increment of
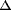 m/z n x 68
from the main metabolite (m/z 236) were characterized in the
corresponding ion chromatograms by HPLC-negative ESI-MS analysis.
Since the repetitive unit in polyisoprene has a molecular mass of 68
Da, it can be safely assumed that the minor products have the same
functional groups as the main metabolite (m/z 236) and that
the only structural difference is the number of isoprene units,
m/z n x 68
from the main metabolite (m/z 236) were characterized in the
corresponding ion chromatograms by HPLC-negative ESI-MS analysis.
Since the repetitive unit in polyisoprene has a molecular mass of 68
Da, it can be safely assumed that the minor products have the same
functional groups as the main metabolite (m/z 236) and that
the only structural difference is the number of isoprene units,
 CH2-C(CH3)
CH2-C(CH3)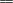 CH-CH2
CH-CH2 ,
incorporated between the terminal functional groups. The concentration
of these oligomers was 1 to 2 orders of magnitude lower than
that of ODTD. ODTD thus apparently is the principal end product of
the RoxA-catalyzed cleavage of polyisoprene. These findings are
consistent with the assumption that RoxA cleaves polyisoprene
oxidatively at regular intervals, cutting off three isoprene units
per step. Our results are in good agreement with previous findings of
Tsuchii and coworkers, who identified a whole range of related
oligomers with more than 100 isoprene units in addition to ODTD . However, these results were obtained with
undefined culture fluid, and it was not known how many enzymes were
involved. Presumably, the concentration and/or activity of RoxA in
the culture fluid in the experiments of Tsuchii et al. was not high
enough to allow complete degradation of polyisoprene.
,
incorporated between the terminal functional groups. The concentration
of these oligomers was 1 to 2 orders of magnitude lower than
that of ODTD. ODTD thus apparently is the principal end product of
the RoxA-catalyzed cleavage of polyisoprene. These findings are
consistent with the assumption that RoxA cleaves polyisoprene
oxidatively at regular intervals, cutting off three isoprene units
per step. Our results are in good agreement with previous findings of
Tsuchii and coworkers, who identified a whole range of related
oligomers with more than 100 isoprene units in addition to ODTD . However, these results were obtained with
undefined culture fluid, and it was not known how many enzymes were
involved. Presumably, the concentration and/or activity of RoxA in
the culture fluid in the experiments of Tsuchii et al. was not high
enough to allow complete degradation of polyisoprene.
Trypsin fingerprint analysis of RoxA confirmed that RoxA
is identical to the product of a recently cloned gene assumed to
be involved in rubber degradation . The presence of a
functional signal sequence in the cloned gene was in agreement with
the extracellular localization of RoxA. Comparison of the amino
acid sequence of RoxA deduced from the gene with the database
revealed the presence of several related amino acid sequences of
hypothetical proteins. In addition to related sequences found
previously , sequences coding for a hypothetical protein
of Pirellula sp. (gi32473529), hypothetical protein Bd3821 of
Bdellovibrio bacteriovorus (gi42525145), and some hypothetical
proteins deduced from sequences of environmental samples were
found. A function has not been identified for any of the related
proteins; however, the RoxA sequence and the most closely related
sequences found in the database contain a conserved sequence motif,
MauG of cytochrome c peroxidases, which is consistent with the
oxidative function of RoxA in Xanthomonas sp. RoxA contained
approximately 2 mol of heme per mol of protein. This result is in
agreement with data for the corresponding gene roxA that
postulate the presence of two covalently bound heme molecules per
molecule of RoxA . Experiments to extract heme
with solvents (acid ethyl acetate or acid methyl ethyl ketone) from
purified RoxA were not successful (unpublished observations),
confirming the covalent binding of heme to the protein. The
inhibition of RoxA by cyanide and carbon monoxide and the shift of
the Soret band (406 nm) upon reduction with dithionite (418 nm) or
upon incubation with synthetic rubber (409 nm) are in agreement with
the involvement of heme in the reaction. Interestingly, addition of
catalase did not inhibit RoxA-catalyzed cleavage of NR, suggesting
that (free) hydrogen peroxide is not involved in the reaction. The
negative results for RoxA in the peroxidase assay are in agreement
with the latter finding. Cleavage of polyisoprene by purified RoxA
was strictly dependent on the presence of molecular oxygen. In
conclusion, RoxA is a novel type of oxygenase. Future experiments
will address the function of heme in the reaction mechanism.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutsche Forschungsgemeinschaft
to D.J.
We gratefully acknowledge J. Armbruster (Universität
Stuttgart-Hohenheim) for assistance with the HPLC-MS and gas
chromatography-MS techniques and for helpful discussions. We also
thank M. Priemer and A. Nordheim (Universität Tübingen) for the
trypsin fingerprint, matrix-assisted laser desorption ionization—time
of flight, and HPLC-MS analyses of RoxA, as well as E. Chua, A. Ikram,
and H. Y. Yeang (Rubber Research Institute of Malaysia) for
providing purified Hevea latex.








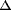 m/z n x 68
from the main metabolite (m/z 236) were characterized in the
corresponding ion chromatograms by HPLC-negative ESI-MS analysis.
Since the repetitive unit in polyisoprene has a molecular mass of 68
Da, it can be safely assumed that the minor products have the same
functional groups as the main metabolite (m/z 236) and that
the only structural difference is the number of isoprene units,
m/z n x 68
from the main metabolite (m/z 236) were characterized in the
corresponding ion chromatograms by HPLC-negative ESI-MS analysis.
Since the repetitive unit in polyisoprene has a molecular mass of 68
Da, it can be safely assumed that the minor products have the same
functional groups as the main metabolite (m/z 236) and that
the only structural difference is the number of isoprene units,